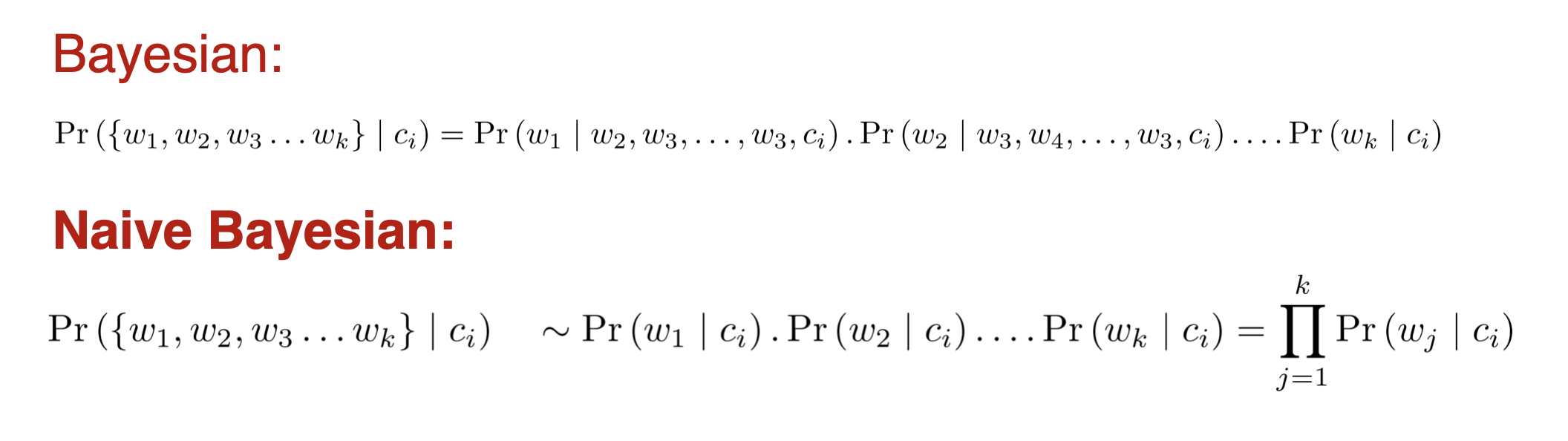Lesson 1.9: Supervised Learning: K-Nearest Neighbours & Naive Bayes
K-Nearest Neighbours
KNN is a simple yet powerful instance-based (or lazy learning) algorithm used for classification and regression. For classification, it assigns a class label to an unlabeled example based on the majority class among its closest neighbors in the training data.
Nearest Neighbor Classification
- Classifies an unlabeled example by assigning it the class of the most similar labeled examples.
- 1-NN: The simplest case where k=1. The label is assigned based on the single closest neighbor.
K-Nearest Neighbors (KNN) Classification
- Generalizes the idea by considering k closest neighbors.
- The class label is determined by majority voting among the k neighbors.
Calculating Distance
KNN relies on distance metrics to determine similarity. Common distance measures include:
Manhattan Distance (L1 Norm)
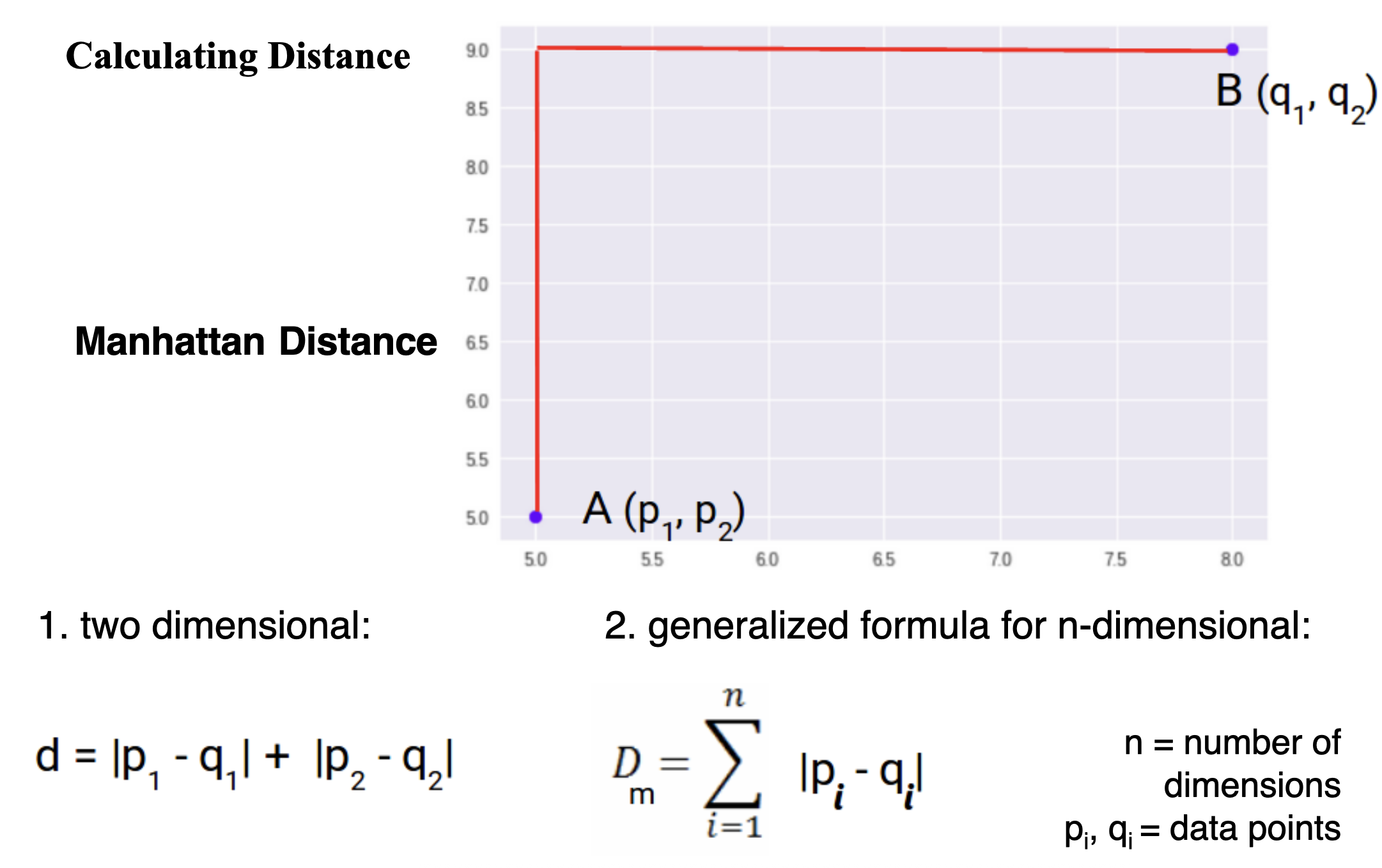
- Sum of absolute differences between coordinates.
Euclidean Distance (L2 Norm)
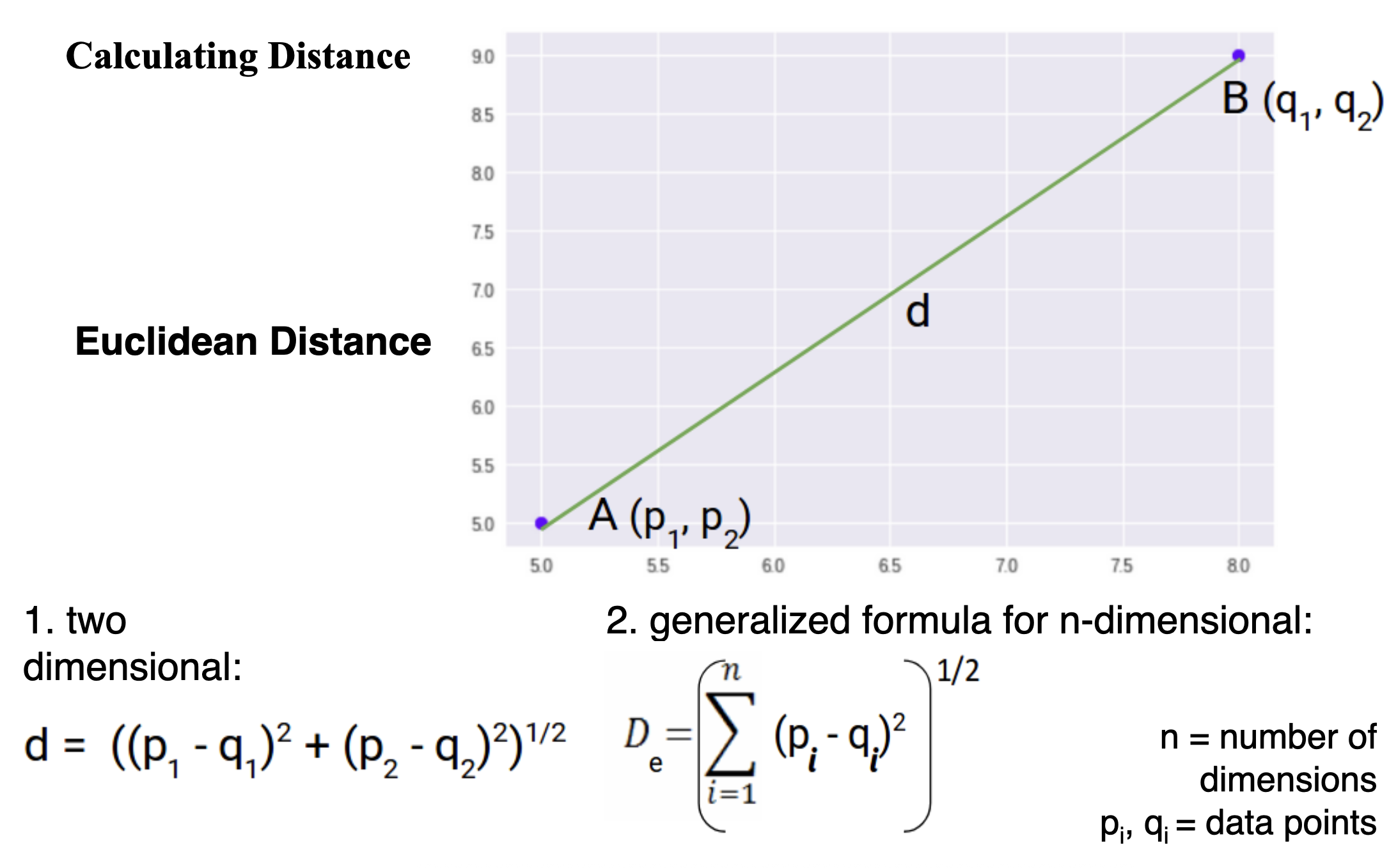
- Straight-line distance between points in space.
Choosing the Right
Small (e.g., )
- Highly sensitive to noise (overfitting).
- Decision boundary is more complex.
Large
- Smoother decision boundaries (underfitting).
- Computationally expensive.
Common Practices for Selecting
- Typically set between 3 and 10.
- A heuristic: where is the number of training samples.
- Deciding how many neighbors to use for KNN determines how well the mode will generalize to future data.
- Choosing a large k reduces the impact or variance caused by noisy data, but can bias the learner such that it runs the risk of ignoring small, but important patterns.
- In practice, choosing k depends on the difficulty of the concept to be learned and the number of records in the training data.
- Typically, k is set somewhere between 3 and 10. One common practice is to set k equal to the square root of the number of training examples.
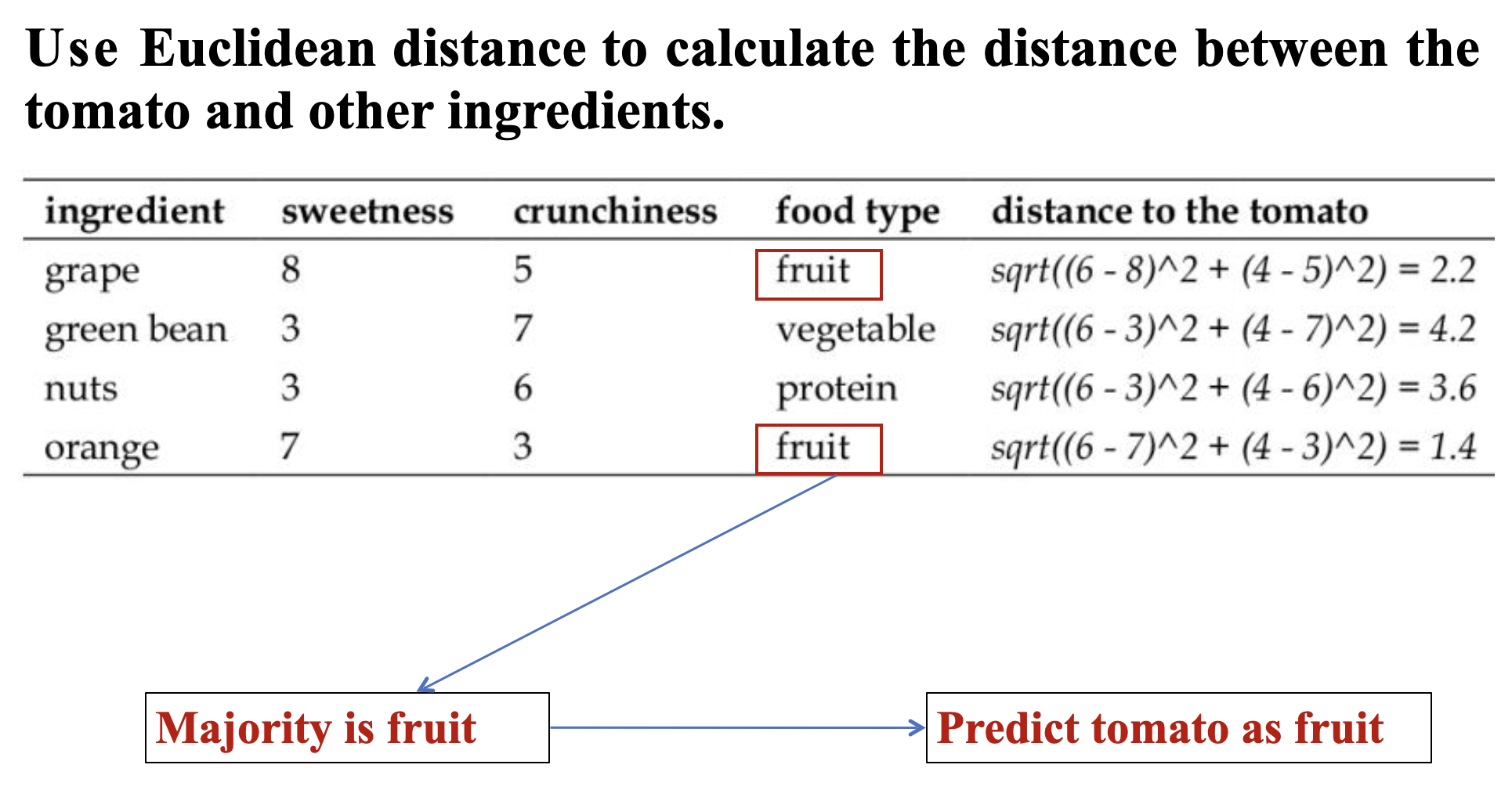
- In the classifier, we might set k = 4, because there were 15 example ingredients in the training data and the square root of 15 is 3.87.
Min-Max Normalization
Since KNN is distance-based, features should be normalized to prevent bias from large-valued features.
- Scales all features to [0,1].
- Ensures equal contribution from all features in distance calculation.
- The traditional method of rescaling features for KNN is min-max normalization.
- This process transforms a feature such that all of its values fall in a range between 0 and 1. The formula for normalizing a feature is as follows. Essentially, the formula subtracts the minimum of feature X from each value and divides by the range of X:
- Normalized feature values can be interpreted as indicating how far, from 0 percent to 100 percent, the original value fell along the range between the original minimum and maximum.
Lazy Learning (Instance-Based Learning)
-
No explicit training phase: Simply stores the training data ("rote learning").
-
Prediction phase is slower: Requires computing distances to all training points for each query.
-
Using the strict definition of learning, a lazy learner is not really learning anything.
-
Instead, it merely stores the training data in it. This allows the training phase to occur very rapidly, with a potential downside being that the process of making predictions tends to be relatively slow.
-
Due to the heavy reliance on the training instances, lazy learning is also known as instance-based learning or rote learning.
-
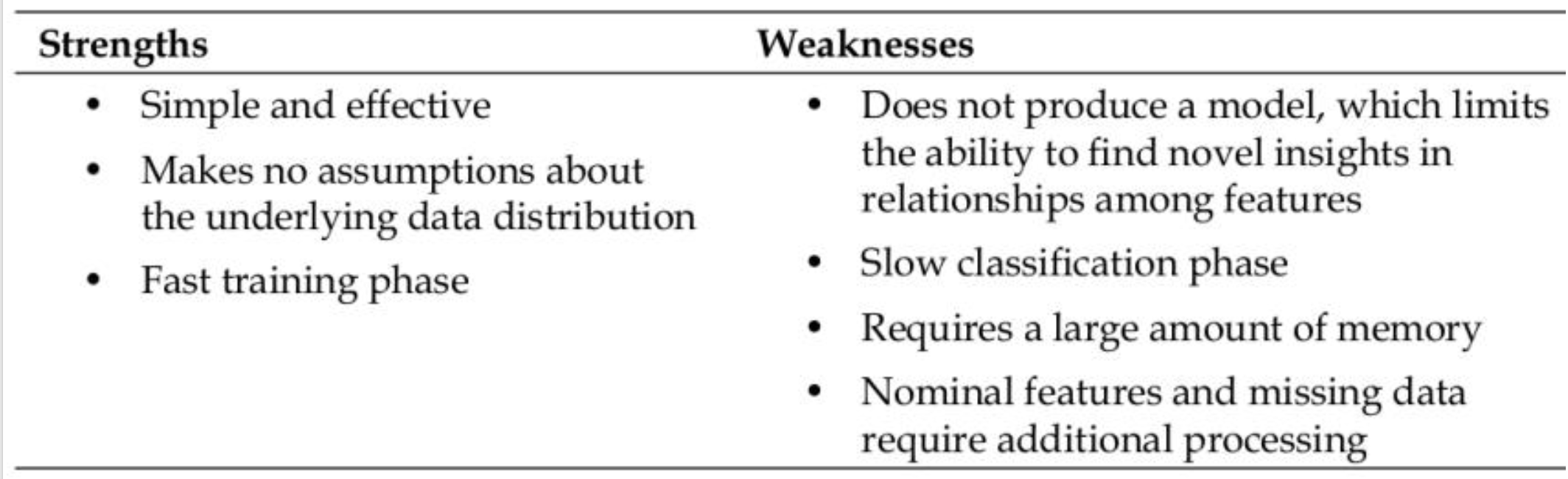
Advantages:
- Adapts immediately to new data.
- No assumptions about data distribution.
-
Disadvantages:
- Computationally expensive for large datasets.
- Sensitive to irrelevant/noisy features.
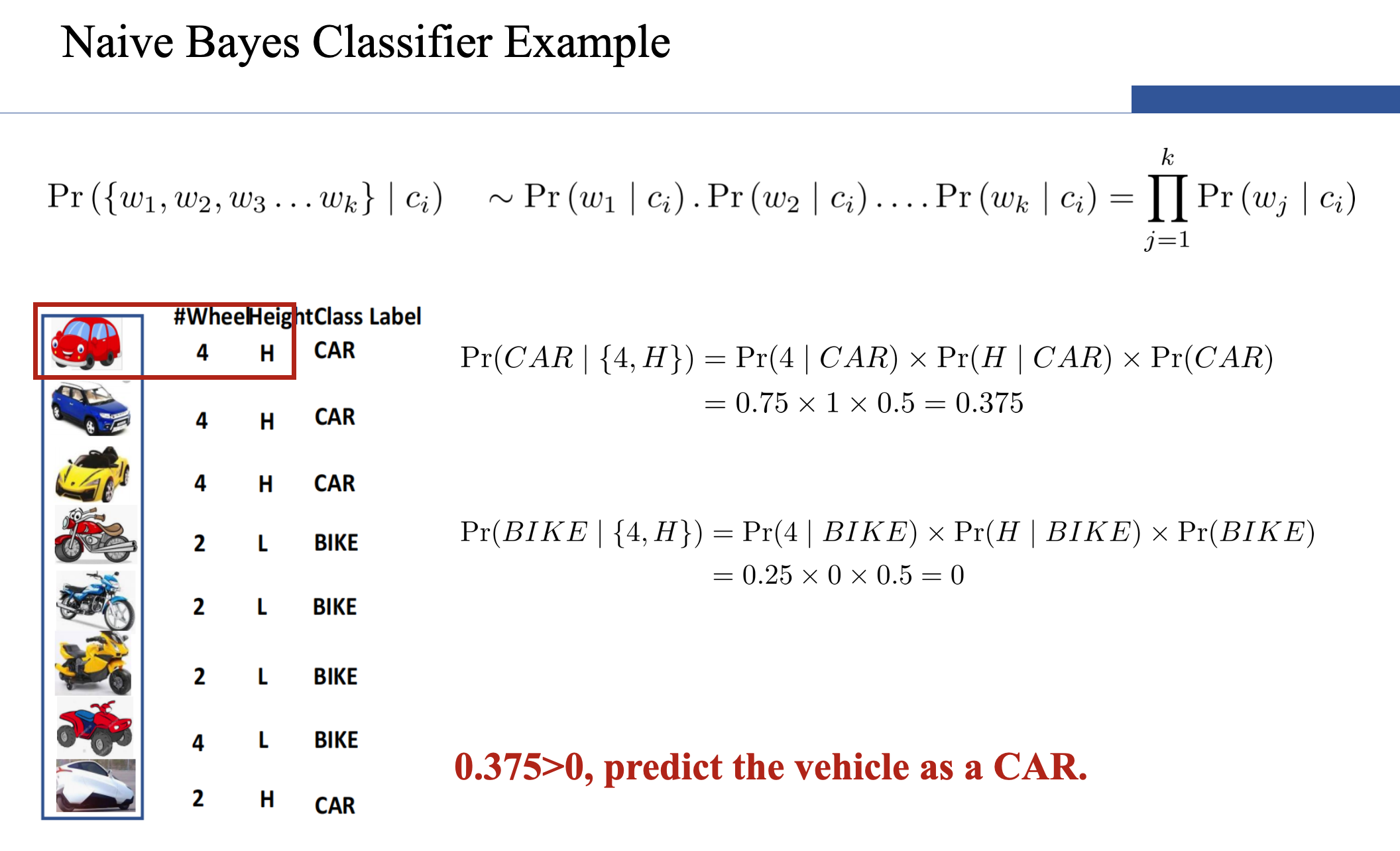
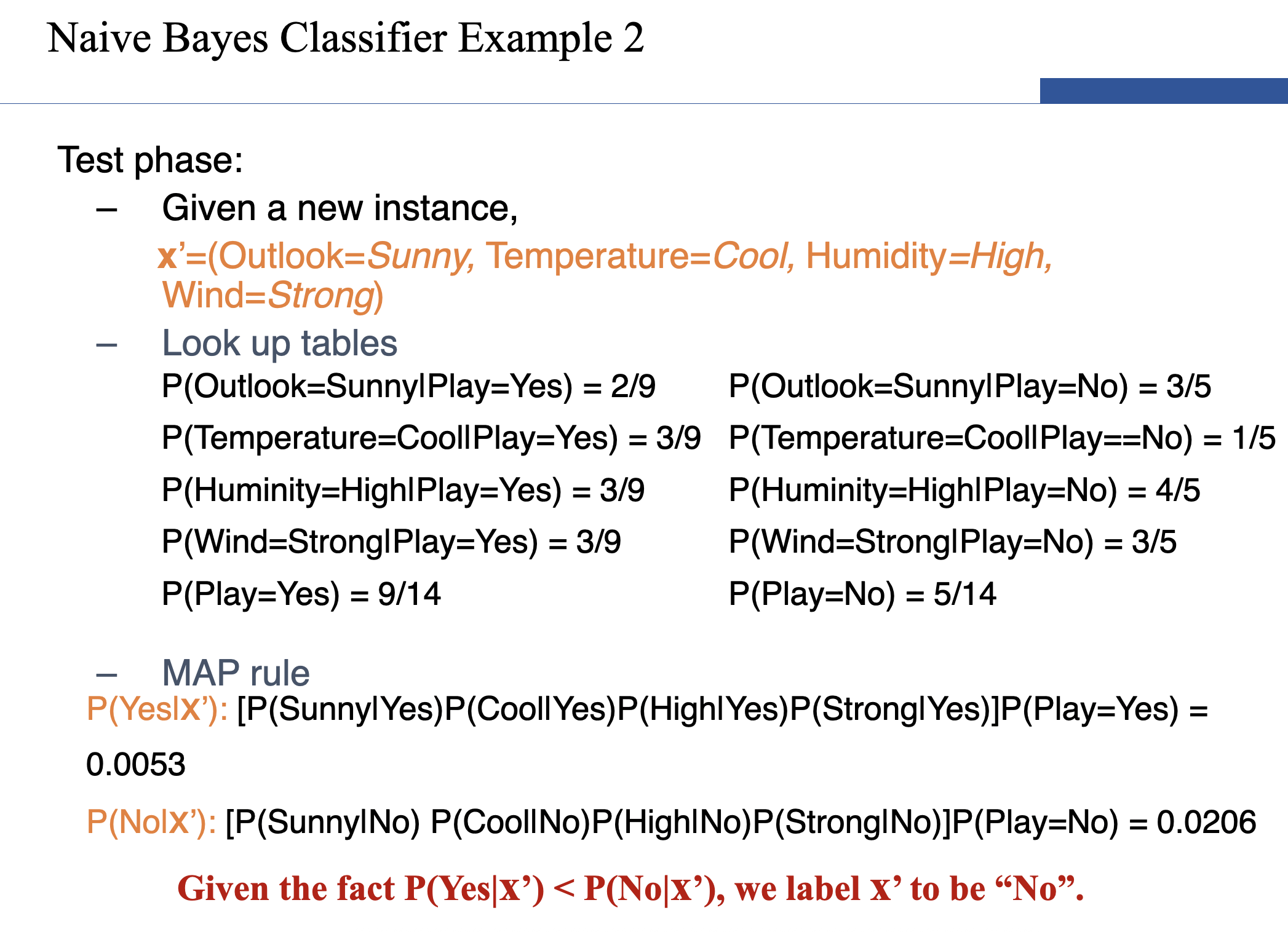
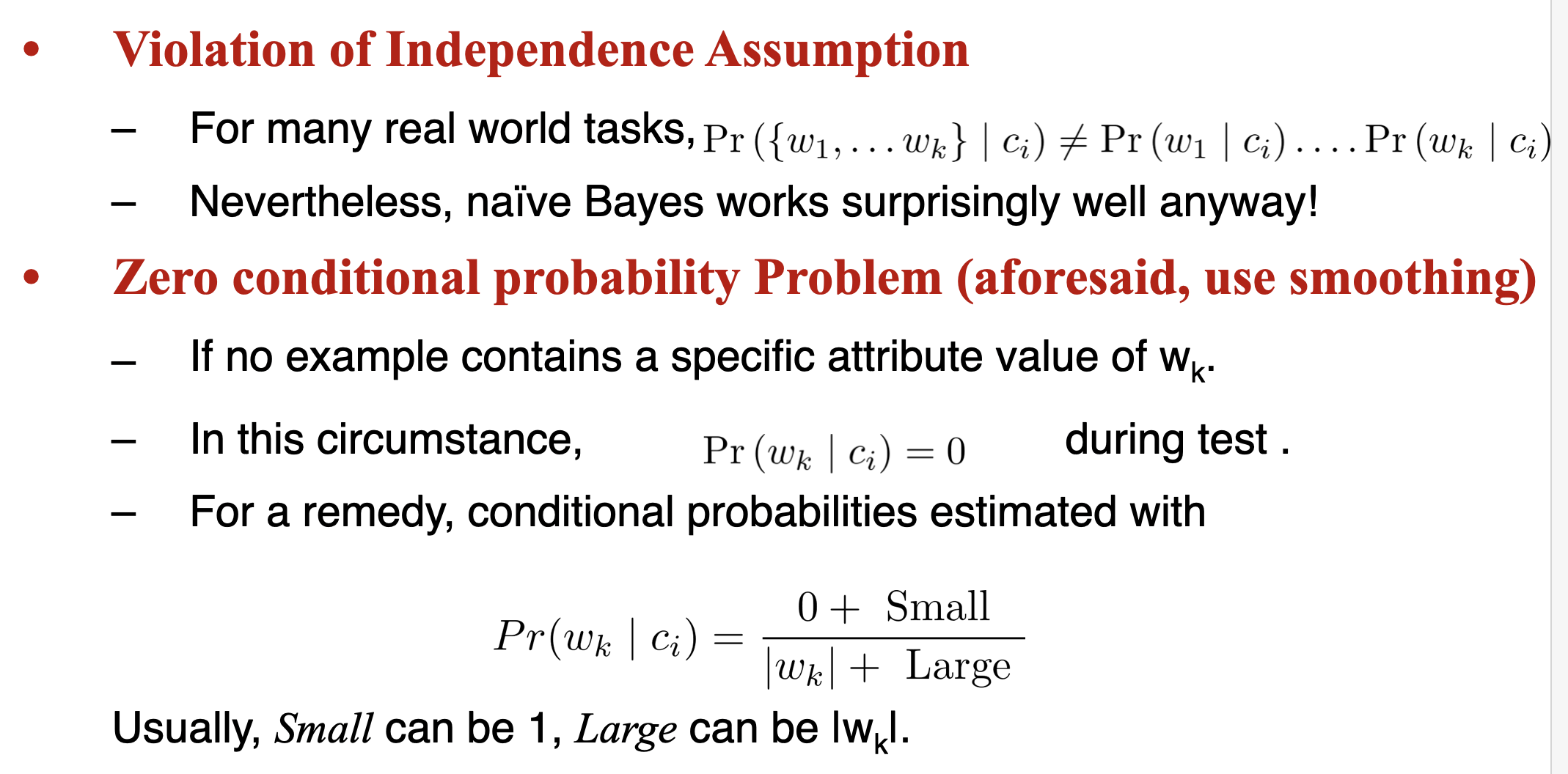
Naive Bayes
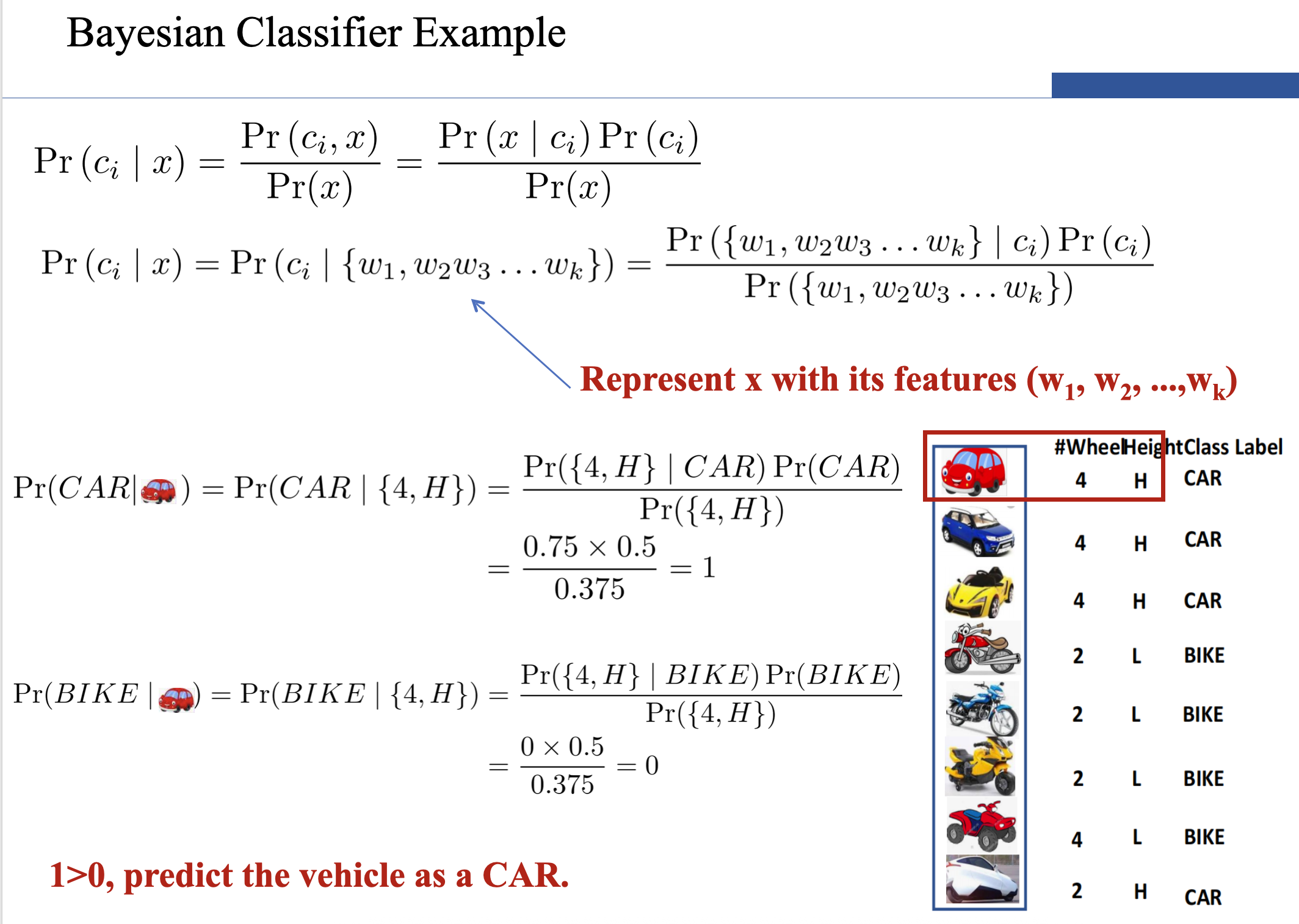
- However, if k (the number of classes) is very large, estimating likelihood is a very expensive task over a large dataset.
- To simplify the estimation, we make an assumption. The features are conditionally independent.